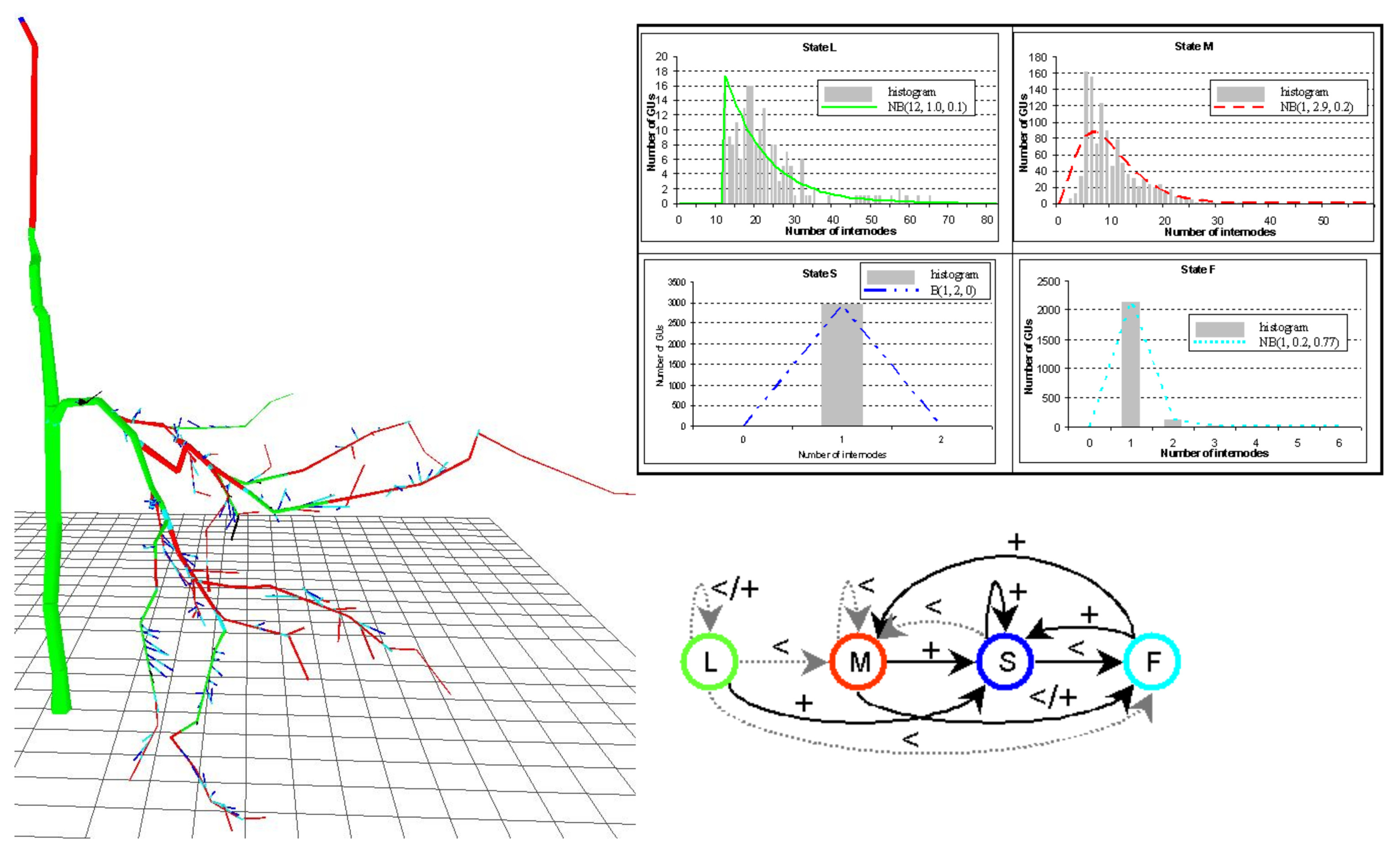
Structure Analysis
Purpose
Structure Analysis is a set of python modules for the analysis of structured data issued from plant phenotyping. It is part of OpenAlean and mainly dedicated to the statistical analysis of graphs, trees and sequences but also includes computational methods for tree matching. Sequence analysis is based on hidden semi-Markov and variable-order Markov chains1. Tree analysis is based on hidden Markov trees2. Tree matching offers possibilities of accounting for different metrics and constraints3. …
Try it !
Docker and singularity images are available at https://gricad-gitlab.univ-grenoble-alpes.fr/durandje/vplants.bak.git (request for access required)
Exemple
An example illustrating sequence analysis with hidden semi-Markov chains (request for access required)
LICENSE
This package is available through the Cecill-C License. A full description of the license can be found here .
Links
- Link to github
References
Guédon, Y., Barthélémy, D., Caraglio, Y., & Costes, E. (2001). Pattern analysis in branching and axillary flowering sequences. Journal of theoretical biology, 212(4), 481-520. ↩︎
Durand, J. B., Guédon, Y., Caraglio, Y., & Costes, E. (2005). Analysis of the plant architecture via tree‐structured statistical models: the hidden Markov tree models. New Phytologist, 166(3), 813-825. ↩︎
Segura, V. , Ouangraoua, A. , Ferraro P., & Costes, E. (2008). Comparison of tree architecture using tree edit distances: application to two-year-old apple tree. Euphytica 161(1-2), 55-164. ↩︎